Abstract
INTRODUCTION: Bone marrow derived mesenchymal stromal cells (BMSC) have been widely used to treat or prevent acute graft-versus-host disease, however the variable efficacies reported between clinical trials suggests considerable heterogeneity between clinical-grade BMSC products. Preincubation of BMSC with IFN-γ may improve their immunosuppressive qualities, reduce the functional diversity of the cell product, and decrease the variability in the clinical results obtained. We aimed to identify the distinct BMSC transcriptome pattern induced by IFN-γ and explored BMSC heterogeneity by studying transcriptional diversity at the single cell level.
METHODS: Human clinical grade BMSC (n=3) were produced at the GMP Clinical Cell Processing Laboratory in the Department of Transfusion Medicine, NIH under an IRB-approved protocol (NCT01071577). BMSC at passage 3, were cultured with or without IFN-γ (10 µg/mL) and were harvested after 12 hours. Single cell 3' digital gene expression profiling for BMSC from three paired conditions (IFN-γ +/-) were performed via the10xTM GemCodeTM Single Cell Platform (10x Genomics, Pleasanton, CA USA) at a targeted cell number of 5000 cells. Single cell RNA-seq library generations, GEM-RT, and cDNA amplification were performed according to manufacturer's instruction and libraries were sequenced on Illumina Hiseq 3000. The Cell Ranger Single-Cell Software was used for sample demultiplexing, barcode processing, single-cell gene counting, and cell clustering.
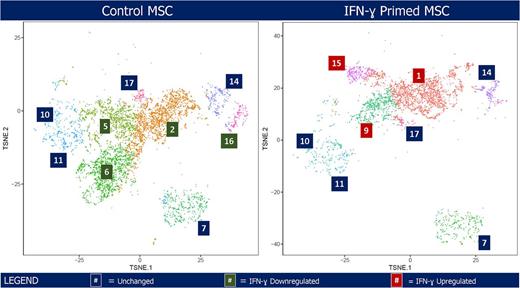
RESULTS: BMSC samples were analyzed for single cell transcriptome at an average of 5188 cells per condition with a mean number of reads of 50,789 per cell and a median of 4,482 genes per cell. Cluster analysis of the six pooled samples revealed clinical grade BMSCs comprised a heterogenous population of at least 17 clusters. Mean count of unique molecular identifiers (UMI) confirmed that all clusters ubiquitously expressed BMSC phenotype markers (CD105, CD73, and CD90). Predominant clusters in control BMSC (clusters 2, 5, and 6) were enriched with distinct expression profiles, indicative of the diversity and heterogeneity of BMSC. Based on Gene Ontology gene set enrichment analyses, cluster 2 was enriched for genes involved in the extracellular matrix, endoplasmic reticulum lumen, and cell adhesion with high expression of IL1β, a proinflammatory cytokine. Clusters 5 and 6 made up populations enriched with a similar set of genes contributing to the mitotic cell cycle checkpoint system, protein serine-threonine kinase activity, and cell division suggesting proliferation potency. Cluster 6 was further characterized by genes involved in DNA replication and ATP binding. IFN-ɣ dramatically altered cluster arrangement, influencing the cell population and its function. For example, major clusters in control BMSC (clusters 2, 5, 6, and 16) significantly diminished in population size and, instead, new clusters 1, 9, and 15 appeared with IFN-ɣ stimulation (Figure). These IFN-ɣ inducible clusters were enriched for genes regulating chemotaxis, leukocyte migration, and MHC class I and II in addition to the type I interferon signaling pathway. Cluster 1 also expressed IDO1, a heme enzyme producing gene responsible for exhibiting immunosuppressive activity. Cluster 9 possessed distinctive features of genes involved in positive regulation of cAMP-mediated signaling and transport vesicle membranes, suggesting extracellular vesicle and exosome activity. Irrespective of IFN-ɣ stimulation, stagnant clusters enriched with G-protein coupled receptor signaling pathway genes remained, representing a vital population in stem cell self-renewal function.
CONCLUSION: Single cell RNA-seq analysis of clinical grade BMSCs revealed a significant heterogeneous population composed of multiple clusters with distinct gene expression patterns. IFN-γ further modified the landscape of transcriptomes of BMSC at a single cell level. This transcriptome diversity may explain the differences in the quality of BMSC products for clinical application. Ongoing experiments include determining the dynamics of the population structure responsible for BMSC diversity and correlating the patterns of single cell transcriptome with the clinical potency of BMSC products.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.